Taxonomy of the bacteria was
historically based on phenotypical physical and chemical characteristics –
phenetic taxonomy. According to "Bergey's Manual of Systematic Bacteriology", all bacteria can be classified into four
divisions, or
phyla according to the constituents of their
cell walls. [
flow chart for classification of obligate intracellular bacteria]
Each division was further subdivided into sections according to -
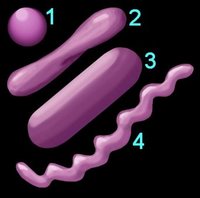
1. Reaction to the
Gram stain due to thick layer of peptidoglycan (Gram +ve) or thin layer of peptidoglycan (Gram -ve). The cell walls of Archaeobacteria contain no peptidoglycan.
2.
Shape (left) – cocci (1), pleomorphic (2), bacillus (3), helical (4) 3.
Arrangement of cells
4.
Oxygen requirement – aerobic, anaerobic, or facultative anaerobe
5.
Motility – flagellated, non-motile
6. Specific
nutritional requirements7.
Trophic (metabolic) properties – autotrophic (chemical or photosynthetic), heterotrophic
The major subdivisions employed in
general taxonomies are:
Domain,
Kingdom,
Phylum,
Class,
Order,
Family,
Genus, and
Species. The mnemonic "Dashing King Philip Came Over From Greater Spain" applies.
These subdivisions may be further subdivided. It is common for bacteria to be subdivided into
Divisions and further subdivided into Orders. The advent of genomics and examination of
16s ribosomal RNA has led to modifications in the classification of the prokaryotes, with the older nomenclatures revised into new classifications of
Phyla.
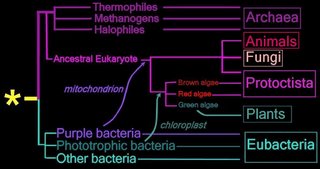
Right - click to enlarge image:
Proposed
endosymbiotic transfer events between the three
Domains and the six
Kingdoms of Life.
(below) Both the
Eubacteria and
Archaea are
prokaryotes, while animals, fungi, plants, and protists are
eukaryotes.
The yellow asterisk
* indicates the
last universal common ancestor (
LUCA), or
universal cenancestor, which is hypothesized as being at the ancestral root of all living organisms. Not the earliest or simplest living organism, and not necessarily the sole example of its type, this organism possessed the genetic material that diverged (about 3.5 Ga) into all current living organisms. A number of terms are employed to refer to the universal cenancestor – last universal ancestor (LUA), last common ancestor (LCA), or last universal common ancestor (LUCA).
Woese and Fox proposed the
Three Domain system:
Eubacteria,
Archaea, and
Eukaryotes.
The Five
Kingdom system was proposed in 1969: Monera (prokaryotes), Protista, Plantae, Fungi, Animalia. Discovery of the
Archaea added the sixth kingdom.
History of taxonomic concepts:
Linnaeus, 1735 – 2 Kingdoms – Animalia, Vegetabilia
Haeckel, 1866 – 3 Kingdoms – Protista, Plantae, Animalia. Image
Haeckel's tree of life.
Chatton, 1937 – 2 Empires – Prokaryota, Eukaryota
Copeland, 1956 – 4 Kingdoms – Monera (
prokaryotes), Protoctista, Plantae, Animalia
Whittaker, 1969 – Monera (
prokaryotes), Fungi, Protista, Plantae, Animalia
Woese et al, 1977 – 6 Kingdom – Eubacteria, Archaea, Protista, Fungi, Plantae, Animalia
Woese and Fox, 1999 – 3 Domain system:
Eubacteria,
Archaea, and
EukaryotesWhen Woese and Fox proposed the 3 Domain system, the term 'Urkaryotes' was proposed for ancestors of eukaryotes prior to their endosymbiotic acquisition of
mitochondria and
chloroplasts from prokaryotes. Margulis and Schwartz proposed that
Kingdom Protista be replaced by Protoctista to reflect inclusion of multicellular organisms that did fit into the other three eukaryotic kingdoms.
"Misunderstanding the Bacteriological Code"The Three Domain system is increasingly accepted. Free Full Text Article :
Phylogenetic structure of the prokaryotic domain: The primary kingdoms. Woese CR,
Fox GE.
Proc Natl Acad Sci U S A. 1977 Nov;74(11):5088-90.
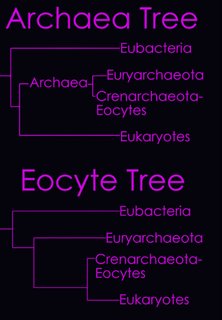
According to the
Tree of Life Web Project, two alternative views on the relationship of the major lineages (omitting viruses) are currently regarded as viable (right - click to enlarge image).

Left - click to enlarge image:
Horizontal gene transfer - gene swapping - has blurred the evolutionary relationships (
image) of prokaryotes, and continues to provide a mechanism for the sharing of antibiotic resistance between bacteria.
See: Trees, vines and nets: Microbial evolution changes its face.Phylogenetic separation into evolutionary relationships (clades), based on comparison of
genomes is likely to supplant phenotypical (phenetic) taxonomies of the prokaryotes.
(
nomenclature)
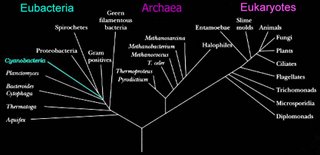
Left - click to enlarge image: Woese has proposed a scheme based on the 16s subunit of
ribosomal RNA, which appears to better illustrate evolutionary relationships within the 3 domains.
Phylogenetic tree of organismsDomain combinations in archaeal, eubacterial and eukaryotic proteomes.
There is a limited repertoire of domain families that are duplicated and combined in different ways to form the set of proteins in a genome. Proteins are gene products, and at the level of genes, duplication, recombination, fusion and fission are the processes that produce new genes. We attempt to gain an overview of these processes by studying the evolutionary units in proteins, domains, in the protein sequences of 40 genomes. The domain and superfamily definitions in the Structural Classification of Proteins Database are used, so that we can view all pairs of adjacent domains in genome sequences in terms of their superfamily combinations. We find 783 out of the 859 superfamilies in SCOP in these genomes, and the 783 families occur in 1307 pairwise combinations. Most families are observed in combination with one or two other families, while a few families are very versatile in their combinatorial behaviour; 209 families do not make combinations with other families. This type of pattern can be described as a scale-free network. We also study the N to C-terminal orientation of domain pairs and domain repeats. The phylogenetic distribution of domain combinations is surveyed, to establish the extent of common and kingdom-specific combinations. Of the kingdom-specific combinations, significantly more combinations consist of families present in all three kingdoms than of families present in one or two kingdoms. Hence, we are led to conclude that recombination between common families, as compared to the invention of new families and recombination among these, has also been a major contribution to the evolution of kingdom-specific and species-specific functions in organisms in all three kingdoms. Finally, we compare the set of the domain combinations in the genomes to those in the RCSB Protein Data Bank, and discuss the implications for structural genomics.
Apic G,
Gough J,
Teichmann SA. Domain combinations in archaeal, eubacterial and eukaryotic proteomes.
J Mol Biol. 2001 Jul 6;310(2):311-25.
Table
Comparisons of Eubacteria, Archaea, and Eukaryotes
Electron acceptors for respiration and methanogenesis in prokaryotes
Glycolysis in bacteria
Lithotrophic prokaryotes
Comparison of plant and bacterial photosynthesis - The Three Domains
View Quicktime Movie - Genetic Data
movie of phylogram construction - image
cladogram - image
Tree of Life— Lateral Gene Transfer Diagram - image
uprooted tree - image
16S ribosomal RNA - image
The "Shrub of Life" - image
A comparison of key characteristics from the three domains of life -
enlarged - Genomics
Animations and Images - Proteins & Proteomics -
Animations and Images – Evolution and Phylogenetics -
Animations and Images - Biodiversity -
Animations and Images – Microbial Diversity –
Animations and Images – Emerging Infectious Diseases -
Animations and Images – HIV & AIDS -
Animations and Images :
Labels: phenetic taxonomy
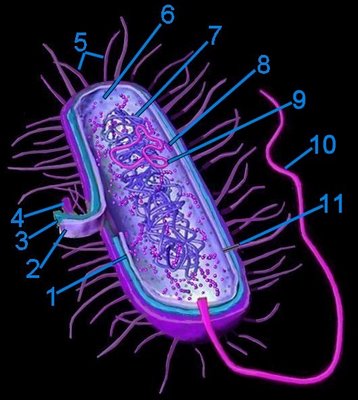 Generalized bacterium
Generalized bacterium











































