Prokaryote structure
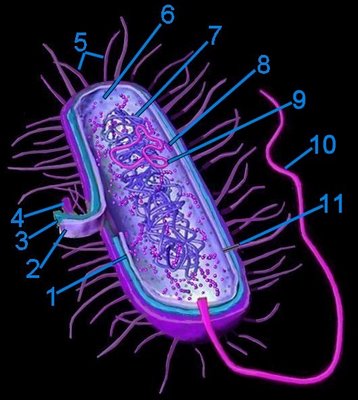 Generalized bacterium
Generalized bacterium1. pericytoplasmic space
2. cytoplasmic membrane
3. cell wall
4. capsule
5. pili
6. cytoplasm
7. cytoplasmic nucleoid
8. 70s ribosomes
9. plasmid
10. flagellum (monotrichous)
11. pore within pilus
Prokaryotes mostly possess one or two* chromosomes termed nucleoids (tem). Because prokaryotes lack a membrane enclosed nucleus their DNA is usually contained in circular structures located within the cytosol, but may be organized as linear strands that are typically attached to the cytoplasmic membrane. Plasmids are small circular, extrachromosomal genetic elements that can be transmitted from one bacterium to another through the pili during conjugation.
*for example, Vibrio cholerae and Deinococcus radiodurans
Cell wall
The Gram stain for bacteria allows differentiation according to thickness of the layer of peptidoglycan (murein, im) in the cell wall. Bacteria that stain heavily (Gram +ve) have a thick monolayer of peptidoglycan compared to the thin or absent layer of peptidoglycan in (bilayer) bacteria that do not take up the stain (Gram -ve). The cell walls of Archaeobacteria contain no peptidoglycan (murein), rather they contain pseudomurein, complex carbohydrates, or protein-glycoproteins.
S-layers comprise one of the most common surface structures on archaea and bacteria. These surface layers have now been identified in hundreds of different species belonging to all major phylogenetic groups of bacteria, and they represent a feature common to almost all archaea (recent compilation 133). S-layers are monomolecular crystalline arrays of proteinaceous subunits (125, 131, 132).
Tables Cell walls of Prokaryotes Comparisons of Eubacteria, Archaea, and Eukaryotes Electron acceptors for respiration and methanogenesis in prokaryotes Glycolysis in bacteria Lithotrophic prokaryotes Structure of bacteriochlorophylls Comparison of plant and bacterial photosynthesis :
Diagrams: Eubacteria : peptidoglycan : gram + gram - peptidoglycan : Gram + : Gram - : P. aeruginosa comp-mod : gram -ve : antimicrobials gram + gram - : cell walls gram + gram - mycobacteria : comparison gram + / gram - cell walls : b-w g+ g- : Archaea : Gram positive archaeal cell wall : Gram negative archaeal cell wall : Unusual cell wall of Deinococcus radiodurans :
: S-layer Freeze etched sem :
The structure of secondary cell wall polymers: how Gram-positive bacteria stick their cell walls together.
The cell wall of Gram-positive bacteria has been a subject of detailed chemical study over the past five decades. Outside the cytoplasmic membrane of these organisms the fundamental polymer is peptidoglycan (PG), which is responsible for the maintenance of cell shape and osmotic stability. In addition, typical essential cell wall polymers such as teichoic or teichuronic acids are linked to some of the peptidoglycan chains. In this review these compounds are considered as 'classical' cell wall polymers. In the course of recent investigations of bacterial cell surface layers (S-layers) a different class of 'non-classical' secondary cell wall polymers (SCWPs) has been identified, which is involved in anchoring of S-layers to the bacterial cell surface. Comparative analyses have shown considerable differences in chemical composition, overall structure and charge behaviour of these SCWPs. This review discusses the progress that has been made in understanding the structural principles of SCWPs, which may have useful applications in S-layer-based 'supramolecular construction kits' in nanobiotechnology.
Schaffer C, Messner P. The structure of secondary cell wall polymers: how Gram-positive bacteria stick their cell walls together. (Free Full Text Article) Microbiology. 2005 Mar;151(Pt 3):643-51.
Molecular organization of selected prokaryotic S-layer proteins.
Regular crystalline surface layers (S-layers) are widespread among prokaryotes and probably represent the earliest cell wall structures. S-layer genes have been found in approximately 400 different species of the prokaryotic domains bacteria and archaea. S-layers usually consist of a single (glyco-)protein species with molecular masses ranging from about 40 to 200 kDa that form lattices of oblique, tetragonal, or hexagonal architecture. The primary sequences of hyperthermophilic archaeal species exhibit some characteristic signatures. Further adaptations to their specific environments occur by various post-translational modifications, such as linkage of glycans, lipids, phosphate, and sulfate groups to the protein or by proteolytic processing. Specific domains direct the anchoring of the S-layer to the underlying cell wall components and transport across the cytoplasma membrane. In addition to their presumptive original role as protective coats in archaea and bacteria, they have adapted new functions, e.g., as molecular sieves, attachment sites for extracellular enzymes, and virulence factors.
Claus H, Akca E, Debaerdemaeker T, Evrard C, Declercq JP, Harris JR, Schlott B, Konig H.
Molecular organization of selected prokaryotic S-layer proteins. Can J Microbiol. 2005 Sep;51(9):731-43.
Glycoproteins in prokaryotes. [Arch Microbiol. 1997] PMID: 9382700
Structural research on surface layers: a focus on stability, surface layer homology domains, and surface layer-cell wall interactions. [J Struct Biol. 1998] PMID: 10049812
Prokaryotic glycosylation. [Proteomics. 2001] PMID: 11680871
Glycobiology of surface layer proteins. [Biochimie. 2001] PMID: 11522387
Stress genes and proteins in the archaea. [Microbiol Mol Biol Rev. 1999] PMID: 10585970
See all Related Articles...
S-Layer proteins.
Cell walls are an important structural component of prokaryotic organisms and essential for many aspects of their life. Particularly, the diverse structures of the outermost boundary layers strongly reflect adaptations of organisms to specific ecological and environmental conditions (6).
Over the past 3 decades of research, it has become apparent that one of the most common surface structures on archaea and bacteria are monomolecular crystalline arrays of proteinaceous subunits termed surface layers or S-layers (125, 131, 132). Since S-layer-carrying organisms are ubiquitous in the biosphere and because S-layers represent one of the most abundant cellular proteins, it is now obvious that these metabolically expensive products must provide the organisms with an advantage of selection in very different habitats (133). This minireview provides a brief survey of the current state of our knowledge about S-layers with a particular focus on molecular biological and genetic aspects. Other recent reviews (5, 7, 127, 133, 135) are recommended for a more detailed introduction to and treatises on this subject.
Sara M, Sleytr UB. S-Layer proteins. (Free Full Text Article) J Bacteriol. 2000 Feb;182(4):859-68.
Prokaryotic glycosylation. [Proteomics. 2001] PMID: 11680871
Common history at the origin of the position-function correlation in transcriptional regulators in archaea and bacteria. [J Mol Evol. 2001] PMID: 11523004
[Homologous protein domains in superkingdoms Archaea, Bacteria, and Eukaryota and the problem of the origin of eukaryotes] [Izv Akad Nauk Ser Biol. 2005] PMID: 16212260
See all Related Articles...
Cell wall polymers in Archaea (Archaebacteria).
The distribution of the various cell wall and cell envelope (S-layer) polymers among the main lineages of the domain Archaea (Archaebacteria) and the chemical composition and primary structure of polymers forming rigid cell wall sacculi is described. Differences between bacteria and archaea in their sensitivity to antibiotics which inhibit cell wall synthesis in bacteria are discussed.
Kandler O, Konig H. Cell wall polymers in Archaea (Archaebacteria). Cell Mol Life Sci. 1998 Apr;54(4):305-8.
beta-Lactamases are absent from Archaea (archaebacteria). [Microb Drug Resist. 1996] PMID: 9158771
Structure of anionic carbohydrate-containing cell wall polymers in several representatives of the order actinomycetales. [Biochemistry (Mosc). 2000] PMID: 11092967
Anionic polymers in cell walls of gram-positive bacteria. [Biochemistry (Mosc). 1997] PMID: 9360295
The response of selected members of the archaea to the gram stain. [Microbiology. 1996] PMID: 8885405
Life's third domain (Archaea): an established fact or an endangered paradigm? [Theor Popul Biol. 1998] PMID: 9733652
See all Related Articles...
S-layers comprise one of the most common surface structures on archaea and bacteria. These surface layers have now been identified in hundreds of different species belonging to all major phylogenetic groups of bacteria, and they represent a feature common to almost all archaea (recent compilation 133). S-layers are monomolecular crystalline arrays of proteinaceous subunits (125, 131, 132).
Tables Cell walls of Prokaryotes Comparisons of Eubacteria, Archaea, and Eukaryotes Electron acceptors for respiration and methanogenesis in prokaryotes Glycolysis in bacteria Lithotrophic prokaryotes Structure of bacteriochlorophylls Comparison of plant and bacterial photosynthesis :
Diagrams: Eubacteria : peptidoglycan : gram + gram - peptidoglycan : Gram + : Gram - : P. aeruginosa comp-mod : gram -ve : antimicrobials gram + gram - : cell walls gram + gram - mycobacteria : comparison gram + / gram - cell walls : b-w g+ g- : Archaea : Gram positive archaeal cell wall : Gram negative archaeal cell wall : Unusual cell wall of Deinococcus radiodurans :
: S-layer Freeze etched sem :
The structure of secondary cell wall polymers: how Gram-positive bacteria stick their cell walls together.
The cell wall of Gram-positive bacteria has been a subject of detailed chemical study over the past five decades. Outside the cytoplasmic membrane of these organisms the fundamental polymer is peptidoglycan (PG), which is responsible for the maintenance of cell shape and osmotic stability. In addition, typical essential cell wall polymers such as teichoic or teichuronic acids are linked to some of the peptidoglycan chains. In this review these compounds are considered as 'classical' cell wall polymers. In the course of recent investigations of bacterial cell surface layers (S-layers) a different class of 'non-classical' secondary cell wall polymers (SCWPs) has been identified, which is involved in anchoring of S-layers to the bacterial cell surface. Comparative analyses have shown considerable differences in chemical composition, overall structure and charge behaviour of these SCWPs. This review discusses the progress that has been made in understanding the structural principles of SCWPs, which may have useful applications in S-layer-based 'supramolecular construction kits' in nanobiotechnology.
Schaffer C, Messner P. The structure of secondary cell wall polymers: how Gram-positive bacteria stick their cell walls together. (Free Full Text Article) Microbiology. 2005 Mar;151(Pt 3):643-51.
Molecular organization of selected prokaryotic S-layer proteins.
Regular crystalline surface layers (S-layers) are widespread among prokaryotes and probably represent the earliest cell wall structures. S-layer genes have been found in approximately 400 different species of the prokaryotic domains bacteria and archaea. S-layers usually consist of a single (glyco-)protein species with molecular masses ranging from about 40 to 200 kDa that form lattices of oblique, tetragonal, or hexagonal architecture. The primary sequences of hyperthermophilic archaeal species exhibit some characteristic signatures. Further adaptations to their specific environments occur by various post-translational modifications, such as linkage of glycans, lipids, phosphate, and sulfate groups to the protein or by proteolytic processing. Specific domains direct the anchoring of the S-layer to the underlying cell wall components and transport across the cytoplasma membrane. In addition to their presumptive original role as protective coats in archaea and bacteria, they have adapted new functions, e.g., as molecular sieves, attachment sites for extracellular enzymes, and virulence factors.
Claus H, Akca E, Debaerdemaeker T, Evrard C, Declercq JP, Harris JR, Schlott B, Konig H.
Molecular organization of selected prokaryotic S-layer proteins. Can J Microbiol. 2005 Sep;51(9):731-43.
Glycoproteins in prokaryotes. [Arch Microbiol. 1997] PMID: 9382700
Structural research on surface layers: a focus on stability, surface layer homology domains, and surface layer-cell wall interactions. [J Struct Biol. 1998] PMID: 10049812
Prokaryotic glycosylation. [Proteomics. 2001] PMID: 11680871
Glycobiology of surface layer proteins. [Biochimie. 2001] PMID: 11522387
Stress genes and proteins in the archaea. [Microbiol Mol Biol Rev. 1999] PMID: 10585970
See all Related Articles...
S-Layer proteins.
Cell walls are an important structural component of prokaryotic organisms and essential for many aspects of their life. Particularly, the diverse structures of the outermost boundary layers strongly reflect adaptations of organisms to specific ecological and environmental conditions (6).
Over the past 3 decades of research, it has become apparent that one of the most common surface structures on archaea and bacteria are monomolecular crystalline arrays of proteinaceous subunits termed surface layers or S-layers (125, 131, 132). Since S-layer-carrying organisms are ubiquitous in the biosphere and because S-layers represent one of the most abundant cellular proteins, it is now obvious that these metabolically expensive products must provide the organisms with an advantage of selection in very different habitats (133). This minireview provides a brief survey of the current state of our knowledge about S-layers with a particular focus on molecular biological and genetic aspects. Other recent reviews (5, 7, 127, 133, 135) are recommended for a more detailed introduction to and treatises on this subject.
Sara M, Sleytr UB. S-Layer proteins. (Free Full Text Article) J Bacteriol. 2000 Feb;182(4):859-68.
Prokaryotic glycosylation. [Proteomics. 2001] PMID: 11680871
Common history at the origin of the position-function correlation in transcriptional regulators in archaea and bacteria. [J Mol Evol. 2001] PMID: 11523004
[Homologous protein domains in superkingdoms Archaea, Bacteria, and Eukaryota and the problem of the origin of eukaryotes] [Izv Akad Nauk Ser Biol. 2005] PMID: 16212260
See all Related Articles...
Cell wall polymers in Archaea (Archaebacteria).
The distribution of the various cell wall and cell envelope (S-layer) polymers among the main lineages of the domain Archaea (Archaebacteria) and the chemical composition and primary structure of polymers forming rigid cell wall sacculi is described. Differences between bacteria and archaea in their sensitivity to antibiotics which inhibit cell wall synthesis in bacteria are discussed.
Kandler O, Konig H. Cell wall polymers in Archaea (Archaebacteria). Cell Mol Life Sci. 1998 Apr;54(4):305-8.
beta-Lactamases are absent from Archaea (archaebacteria). [Microb Drug Resist. 1996] PMID: 9158771
Structure of anionic carbohydrate-containing cell wall polymers in several representatives of the order actinomycetales. [Biochemistry (Mosc). 2000] PMID: 11092967
Anionic polymers in cell walls of gram-positive bacteria. [Biochemistry (Mosc). 1997] PMID: 9360295
The response of selected members of the archaea to the gram stain. [Microbiology. 1996] PMID: 8885405
Life's third domain (Archaea): an established fact or an endangered paradigm? [Theor Popul Biol. 1998] PMID: 9733652
See all Related Articles...
Labels: Archaea, cell walls, Eubacteria, glycoproteins, Gram stain, peptidoglycan, PG, pseudomurein, S-layers, SCWP, secondary cell wall polymers, teichoic, teichuronic
Taxonomy and Phylogeny
Taxonomy of the bacteria was historically based on phenotypical physical and chemical characteristics – phenetic taxonomy. According to "Bergey's Manual of Systematic Bacteriology", all bacteria can be classified into four divisions, or phyla according to the constituents of their cell walls. [flow chart for classification of obligate intracellular bacteria]
Each division was further subdivided into sections according to -
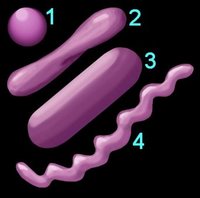 1. Reaction to the Gram stain due to thick layer of peptidoglycan (Gram +ve) or thin layer of peptidoglycan (Gram -ve). The cell walls of Archaeobacteria contain no peptidoglycan.
1. Reaction to the Gram stain due to thick layer of peptidoglycan (Gram +ve) or thin layer of peptidoglycan (Gram -ve). The cell walls of Archaeobacteria contain no peptidoglycan.
2. Shape (left) – cocci (1), pleomorphic (2), bacillus (3), helical (4) 3. Arrangement of cells
4. Oxygen requirement – aerobic, anaerobic, or facultative anaerobe
5. Motility – flagellated, non-motile
6. Specific nutritional requirements
7. Trophic (metabolic) properties – autotrophic (chemical or photosynthetic), heterotrophic
The major subdivisions employed in general taxonomies are:
Domain, Kingdom, Phylum, Class, Order, Family, Genus, and Species. The mnemonic "Dashing King Philip Came Over From Greater Spain" applies.
These subdivisions may be further subdivided. It is common for bacteria to be subdivided into Divisions and further subdivided into Orders. The advent of genomics and examination of 16s ribosomal RNA has led to modifications in the classification of the prokaryotes, with the older nomenclatures revised into new classifications of Phyla.
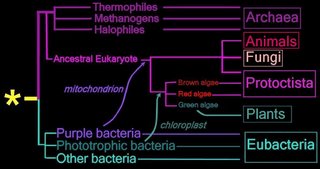 Right - click to enlarge image: Proposed endosymbiotic transfer events between the three Domains and the six Kingdoms of Life. (below) Both the Eubacteria and Archaea are prokaryotes, while animals, fungi, plants, and protists are eukaryotes.
Right - click to enlarge image: Proposed endosymbiotic transfer events between the three Domains and the six Kingdoms of Life. (below) Both the Eubacteria and Archaea are prokaryotes, while animals, fungi, plants, and protists are eukaryotes.
The yellow asterisk * indicates the last universal common ancestor (LUCA), or universal cenancestor, which is hypothesized as being at the ancestral root of all living organisms. Not the earliest or simplest living organism, and not necessarily the sole example of its type, this organism possessed the genetic material that diverged (about 3.5 Ga) into all current living organisms. A number of terms are employed to refer to the universal cenancestor – last universal ancestor (LUA), last common ancestor (LCA), or last universal common ancestor (LUCA).
Woese and Fox proposed the Three Domain system: Eubacteria, Archaea, and Eukaryotes.
The Five Kingdom system was proposed in 1969: Monera (prokaryotes), Protista, Plantae, Fungi, Animalia. Discovery of the Archaea added the sixth kingdom.
History of taxonomic concepts:
Linnaeus, 1735 – 2 Kingdoms – Animalia, Vegetabilia
Haeckel, 1866 – 3 Kingdoms – Protista, Plantae, Animalia. Image Haeckel's tree of life.
Chatton, 1937 – 2 Empires – Prokaryota, Eukaryota
Copeland, 1956 – 4 Kingdoms – Monera (prokaryotes), Protoctista, Plantae, Animalia
Whittaker, 1969 – Monera (prokaryotes), Fungi, Protista, Plantae, Animalia
Woese et al, 1977 – 6 Kingdom – Eubacteria, Archaea, Protista, Fungi, Plantae, Animalia
Woese and Fox, 1999 – 3 Domain system: Eubacteria, Archaea, and Eukaryotes
When Woese and Fox proposed the 3 Domain system, the term 'Urkaryotes' was proposed for ancestors of eukaryotes prior to their endosymbiotic acquisition of mitochondria and chloroplasts from prokaryotes. Margulis and Schwartz proposed that Kingdom Protista be replaced by Protoctista to reflect inclusion of multicellular organisms that did fit into the other three eukaryotic kingdoms.
"Misunderstanding the Bacteriological Code"
The Three Domain system is increasingly accepted. Free Full Text Article : Phylogenetic structure of the prokaryotic domain: The primary kingdoms. Woese CR, Fox GE. Proc Natl Acad Sci U S A. 1977 Nov;74(11):5088-90.
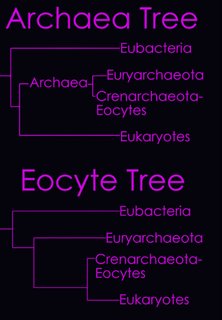
According to the Tree of Life Web Project, two alternative views on the relationship of the major lineages (omitting viruses) are currently regarded as viable (right - click to enlarge image).
 Left - click to enlarge image: Horizontal gene transfer - gene swapping - has blurred the evolutionary relationships (image) of prokaryotes, and continues to provide a mechanism for the sharing of antibiotic resistance between bacteria. See: Trees, vines and nets: Microbial evolution changes its face.
Left - click to enlarge image: Horizontal gene transfer - gene swapping - has blurred the evolutionary relationships (image) of prokaryotes, and continues to provide a mechanism for the sharing of antibiotic resistance between bacteria. See: Trees, vines and nets: Microbial evolution changes its face.
Phylogenetic separation into evolutionary relationships (clades), based on comparison of genomes is likely to supplant phenotypical (phenetic) taxonomies of the prokaryotes.
(nomenclature)
 Left - click to enlarge image: Woese has proposed a scheme based on the 16s subunit of ribosomal RNA, which appears to better illustrate evolutionary relationships within the 3 domains.
Left - click to enlarge image: Woese has proposed a scheme based on the 16s subunit of ribosomal RNA, which appears to better illustrate evolutionary relationships within the 3 domains.
Phylogenetic tree of organisms
Domain combinations in archaeal, eubacterial and eukaryotic proteomes.
There is a limited repertoire of domain families that are duplicated and combined in different ways to form the set of proteins in a genome. Proteins are gene products, and at the level of genes, duplication, recombination, fusion and fission are the processes that produce new genes. We attempt to gain an overview of these processes by studying the evolutionary units in proteins, domains, in the protein sequences of 40 genomes. The domain and superfamily definitions in the Structural Classification of Proteins Database are used, so that we can view all pairs of adjacent domains in genome sequences in terms of their superfamily combinations. We find 783 out of the 859 superfamilies in SCOP in these genomes, and the 783 families occur in 1307 pairwise combinations. Most families are observed in combination with one or two other families, while a few families are very versatile in their combinatorial behaviour; 209 families do not make combinations with other families. This type of pattern can be described as a scale-free network. We also study the N to C-terminal orientation of domain pairs and domain repeats. The phylogenetic distribution of domain combinations is surveyed, to establish the extent of common and kingdom-specific combinations. Of the kingdom-specific combinations, significantly more combinations consist of families present in all three kingdoms than of families present in one or two kingdoms. Hence, we are led to conclude that recombination between common families, as compared to the invention of new families and recombination among these, has also been a major contribution to the evolution of kingdom-specific and species-specific functions in organisms in all three kingdoms. Finally, we compare the set of the domain combinations in the genomes to those in the RCSB Protein Data Bank, and discuss the implications for structural genomics.
Apic G, Gough J, Teichmann SA. Domain combinations in archaeal, eubacterial and eukaryotic proteomes. J Mol Biol. 2001 Jul 6;310(2):311-25.
Table Comparisons of Eubacteria, Archaea, and Eukaryotes Electron acceptors for respiration and methanogenesis in prokaryotes Glycolysis in bacteria Lithotrophic prokaryotes Comparison of plant and bacterial photosynthesis - The Three Domains View Quicktime Movie - Genetic Data movie of phylogram construction - image cladogram - image Tree of Life— Lateral Gene Transfer Diagram - image uprooted tree - image 16S ribosomal RNA - image The "Shrub of Life" - image A comparison of key characteristics from the three domains of life - enlarged - Genomics Animations and Images - Proteins & Proteomics - Animations and Images – Evolution and Phylogenetics - Animations and Images - Biodiversity - Animations and Images – Microbial Diversity – Animations and Images – Emerging Infectious Diseases - Animations and Images – HIV & AIDS - Animations and Images :
Each division was further subdivided into sections according to -
 1. Reaction to the Gram stain due to thick layer of peptidoglycan (Gram +ve) or thin layer of peptidoglycan (Gram -ve). The cell walls of Archaeobacteria contain no peptidoglycan.
1. Reaction to the Gram stain due to thick layer of peptidoglycan (Gram +ve) or thin layer of peptidoglycan (Gram -ve). The cell walls of Archaeobacteria contain no peptidoglycan.2. Shape (left) – cocci (1), pleomorphic (2), bacillus (3), helical (4) 3. Arrangement of cells
4. Oxygen requirement – aerobic, anaerobic, or facultative anaerobe
5. Motility – flagellated, non-motile
6. Specific nutritional requirements
7. Trophic (metabolic) properties – autotrophic (chemical or photosynthetic), heterotrophic
The major subdivisions employed in general taxonomies are:
Domain, Kingdom, Phylum, Class, Order, Family, Genus, and Species. The mnemonic "Dashing King Philip Came Over From Greater Spain" applies.
These subdivisions may be further subdivided. It is common for bacteria to be subdivided into Divisions and further subdivided into Orders. The advent of genomics and examination of 16s ribosomal RNA has led to modifications in the classification of the prokaryotes, with the older nomenclatures revised into new classifications of Phyla.
 Right - click to enlarge image: Proposed endosymbiotic transfer events between the three Domains and the six Kingdoms of Life. (below) Both the Eubacteria and Archaea are prokaryotes, while animals, fungi, plants, and protists are eukaryotes.
Right - click to enlarge image: Proposed endosymbiotic transfer events between the three Domains and the six Kingdoms of Life. (below) Both the Eubacteria and Archaea are prokaryotes, while animals, fungi, plants, and protists are eukaryotes.The yellow asterisk * indicates the last universal common ancestor (LUCA), or universal cenancestor, which is hypothesized as being at the ancestral root of all living organisms. Not the earliest or simplest living organism, and not necessarily the sole example of its type, this organism possessed the genetic material that diverged (about 3.5 Ga) into all current living organisms. A number of terms are employed to refer to the universal cenancestor – last universal ancestor (LUA), last common ancestor (LCA), or last universal common ancestor (LUCA).
Woese and Fox proposed the Three Domain system: Eubacteria, Archaea, and Eukaryotes.
The Five Kingdom system was proposed in 1969: Monera (prokaryotes), Protista, Plantae, Fungi, Animalia. Discovery of the Archaea added the sixth kingdom.
History of taxonomic concepts:
Linnaeus, 1735 – 2 Kingdoms – Animalia, Vegetabilia
Haeckel, 1866 – 3 Kingdoms – Protista, Plantae, Animalia. Image Haeckel's tree of life.
Chatton, 1937 – 2 Empires – Prokaryota, Eukaryota
Copeland, 1956 – 4 Kingdoms – Monera (prokaryotes), Protoctista, Plantae, Animalia
Whittaker, 1969 – Monera (prokaryotes), Fungi, Protista, Plantae, Animalia
Woese et al, 1977 – 6 Kingdom – Eubacteria, Archaea, Protista, Fungi, Plantae, Animalia
Woese and Fox, 1999 – 3 Domain system: Eubacteria, Archaea, and Eukaryotes
When Woese and Fox proposed the 3 Domain system, the term 'Urkaryotes' was proposed for ancestors of eukaryotes prior to their endosymbiotic acquisition of mitochondria and chloroplasts from prokaryotes. Margulis and Schwartz proposed that Kingdom Protista be replaced by Protoctista to reflect inclusion of multicellular organisms that did fit into the other three eukaryotic kingdoms.
"Misunderstanding the Bacteriological Code"
The Three Domain system is increasingly accepted. Free Full Text Article : Phylogenetic structure of the prokaryotic domain: The primary kingdoms. Woese CR, Fox GE. Proc Natl Acad Sci U S A. 1977 Nov;74(11):5088-90.

According to the Tree of Life Web Project, two alternative views on the relationship of the major lineages (omitting viruses) are currently regarded as viable (right - click to enlarge image).
 Left - click to enlarge image: Horizontal gene transfer - gene swapping - has blurred the evolutionary relationships (image) of prokaryotes, and continues to provide a mechanism for the sharing of antibiotic resistance between bacteria. See: Trees, vines and nets: Microbial evolution changes its face.
Left - click to enlarge image: Horizontal gene transfer - gene swapping - has blurred the evolutionary relationships (image) of prokaryotes, and continues to provide a mechanism for the sharing of antibiotic resistance between bacteria. See: Trees, vines and nets: Microbial evolution changes its face.Phylogenetic separation into evolutionary relationships (clades), based on comparison of genomes is likely to supplant phenotypical (phenetic) taxonomies of the prokaryotes.
(nomenclature)
 Left - click to enlarge image: Woese has proposed a scheme based on the 16s subunit of ribosomal RNA, which appears to better illustrate evolutionary relationships within the 3 domains.
Left - click to enlarge image: Woese has proposed a scheme based on the 16s subunit of ribosomal RNA, which appears to better illustrate evolutionary relationships within the 3 domains.Phylogenetic tree of organisms
Domain combinations in archaeal, eubacterial and eukaryotic proteomes.
There is a limited repertoire of domain families that are duplicated and combined in different ways to form the set of proteins in a genome. Proteins are gene products, and at the level of genes, duplication, recombination, fusion and fission are the processes that produce new genes. We attempt to gain an overview of these processes by studying the evolutionary units in proteins, domains, in the protein sequences of 40 genomes. The domain and superfamily definitions in the Structural Classification of Proteins Database are used, so that we can view all pairs of adjacent domains in genome sequences in terms of their superfamily combinations. We find 783 out of the 859 superfamilies in SCOP in these genomes, and the 783 families occur in 1307 pairwise combinations. Most families are observed in combination with one or two other families, while a few families are very versatile in their combinatorial behaviour; 209 families do not make combinations with other families. This type of pattern can be described as a scale-free network. We also study the N to C-terminal orientation of domain pairs and domain repeats. The phylogenetic distribution of domain combinations is surveyed, to establish the extent of common and kingdom-specific combinations. Of the kingdom-specific combinations, significantly more combinations consist of families present in all three kingdoms than of families present in one or two kingdoms. Hence, we are led to conclude that recombination between common families, as compared to the invention of new families and recombination among these, has also been a major contribution to the evolution of kingdom-specific and species-specific functions in organisms in all three kingdoms. Finally, we compare the set of the domain combinations in the genomes to those in the RCSB Protein Data Bank, and discuss the implications for structural genomics.
Apic G, Gough J, Teichmann SA. Domain combinations in archaeal, eubacterial and eukaryotic proteomes. J Mol Biol. 2001 Jul 6;310(2):311-25.
Table Comparisons of Eubacteria, Archaea, and Eukaryotes Electron acceptors for respiration and methanogenesis in prokaryotes Glycolysis in bacteria Lithotrophic prokaryotes Comparison of plant and bacterial photosynthesis - The Three Domains View Quicktime Movie - Genetic Data movie of phylogram construction - image cladogram - image Tree of Life— Lateral Gene Transfer Diagram - image uprooted tree - image 16S ribosomal RNA - image The "Shrub of Life" - image A comparison of key characteristics from the three domains of life - enlarged - Genomics Animations and Images - Proteins & Proteomics - Animations and Images – Evolution and Phylogenetics - Animations and Images - Biodiversity - Animations and Images – Microbial Diversity – Animations and Images – Emerging Infectious Diseases - Animations and Images – HIV & AIDS - Animations and Images :
Labels: phenetic taxonomy
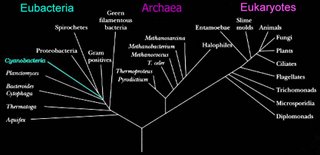
Eubacteria
 The two prokaryotic domains are Eubacteria (bacteria) and Archaea. The Eubacteria are usually called the Bacteria, and the Archaea are sometimes called the Archaebacteria.
The two prokaryotic domains are Eubacteria (bacteria) and Archaea. The Eubacteria are usually called the Bacteria, and the Archaea are sometimes called the Archaebacteria.In the schematic (left) phylogenetic relationships of the bacteria are shown. Successive phyla of bacteria branched off the Bacterial line – beginning with Aquifex, and ending with the Spirochetes.
Schematic based on Woese's Tree of Life, which is based upon studies of 16s ribosomal RNA. (right)
In approximate order of evolutionary divergence with earliest (oldest) at top. Click on group to visit description:
Aquifex
Thermotoga
Green nonsulfur
Deinococcus
Bacteroides-Flavobacteria
Planctomyces
Gram-positive
Chlamydia
Cyanobacteria
Spirochetes
Green sulfur
Proteobacteria (purple)
The Archaea
The Archaea (formerly Archaeobacteria, or Mendosicutes) constitute a recently recognized phylogenetic domain. While Eubacteria and Archaea are similar in structure, they have a different metabolism and genotype. A defining physiological characteristic of Archaea is their ability to live in extreme environments. Thus, these organisms are called “extremophiles” and, unlike Eubacteria and Eukarya, they depend for survival on environmental conditions such as high salinity, extremes of temperature, unusual chemical substrates, or high pressure.
Archaea can be distinguished from bacteria in that their cell walls do not have murein—a peptidoglycan-containing muramic acid. Their plasma membranes are ~4-5 nm in thickness, and are unique in including isopranyl ether lipids – branched isoprenoid hydrocarbons that are linked to glycerol by di-/tetraether bonds. The glycerol diethers are associated with bilayer membranes, whereas the glycerol tetraethers are associated with monolayer membranes. Archaea with mixtures of di- & tetraethers have mixed mono- & bilayer- membranes. Hyperthermophiles have increased membrane fluidity due to branched glycerol tetraethers, and often lack a cell wall. Psychrophiles have lipids comprising mainly unsaturated fatty acids. The plasma membrane of Archaea also contains sulfolipids.
The cell walls of most Archaea comprise a multilayered sacculus (sheet) that surrounds the plasma membrane and provides resistance to osmotic shock. This enables the Archaea (except Thermoplasma) to maintain high intracellular osmotic pressures (15-20 atm). The cells wall may comprise peptidoglycans (Methanobacterium) in which the chief sugars are N-acetyl talosaminouronic acid (NAT or T) and GlcNAc (NAG or G). These are linked to each other by lysozime insensitive (b1->3) glycosidic bond. The T units are linked to L-aminoacid containing oligopeptides by a penicillin-insensitive transpeptidase. In other genera, the cell wall may comprise only non-sulfated polysaccharides (Methanosarcina), only sulfated polysaccharides (Halococcus), only proteins (Methanomicrobium and Methanococcus), or negatively charged aminoacid-containing peptides (Halobacteria).
Images: tem -thermophilic archaeobacteria, line-up

Phylogenetic taxonomy of the Archaea (right), based on 16s rRNA trees.
The Bacteria (eubacteria) were the first prokaryotes recognized. (see phylogeny)
In 1977, Woese and Fox discovered the Archaea based upon their phylogenetic relationships to the Bacteria and the Eucarya. The yellow asterisk indicates the progenote, and the junction between teal and purple lines, the Last Common Ancestor, or Cenancestor.
Woese, Free Full Text Article: Toward a natural system of organisms
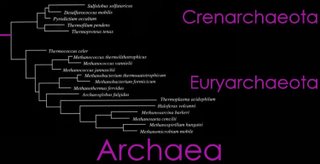 (left) The Domain Archaea is divided into three Phyla: Crenarchaeota, Euryarchaeota, and Nanoarchaeota. A brief phylogenetic relationship of Crenarchaeota and Euryarchaeota are shown in the tree diagram (top right), and a more detailed cladogram is shown below. (click to enlarge)
(left) The Domain Archaea is divided into three Phyla: Crenarchaeota, Euryarchaeota, and Nanoarchaeota. A brief phylogenetic relationship of Crenarchaeota and Euryarchaeota are shown in the tree diagram (top right), and a more detailed cladogram is shown below. (click to enlarge)
From the Taxonomicon, based on Systema Naturae, 2000:
Domain Archaea (Woese, Kandler & Wheelis, 1990)
Phylum Crenarchaeota
Class Thermoprotei
Order Thermoproteales™ Zillig & Stetter, 1982
Order Desulfurococcales
Order Sulfolobales Stetter, 1989
Phylum Euryarchaeota (Woese, Kandler & Wheelis, 1990)
Class Methanobacteria
Class Methanococci
Class Halobacteria
Class Thermoplasmata
Class Thermococci
Class Archaeoglobi Garrity & Holt, 2002
Class Methanopyri
Phylum Nanoarchaeota Huber et al., 2002
Genus Nanoarchaeum™ Huber et al., 2002
The Archaea: A Personal Overview of the Formative Years: "The simple method of growing methanogenic cells in an anaerobic pressurized atmosphere of hydrogen and carbon dioxide was pivotal to the discovery of the Archaea, for hydrogen and carbon dioxide could be replenished with safe containment of high levels of 32P and without exposure of cells to oxygen or contamination.
In November 1977, Woese and Fox proposed that ribosomal RNA sequence characterization could be used to define three "aboriginal lines of descent" (Woese and Fox, 1977). One line, the typical bacteria, was designated "eubacteria." "archaebacteria" was proposed as a name for the methanogen line, and the term "urkaryotes" was proposed for the cytoplasmic component of eukaryotic cells. So the methanogens were Archaebacteria. The name Archaebacteria was suggested by David Nanney. In a way, the term was unfortunate because the case was being made that the methanogens were "old" (i.e., represented a very ancient divergence in evolution), so that they were no more related to bacteria than to eukaryotes. On the other hand, they were bacteria. (Over a decade later, Woese, Kandler, and Wheelis would propose the name "Archaea" for the Archaebacteria [Woese et al., 1990]. By that time, much evidence had accumulated showing the Archaea clearly belonged on the eukaryotic line of descent, and it would be less confusing if the word "bacteria" in Archaebacteria was deleted.)"
more detail: Physiology of the Archaea :
Free Full Text Article : Phylogenetic structure of the prokaryotic domain: The primary kingdoms. Woese CR, Fox GE. Proc Natl Acad Sci U S A. 1977 Nov;74(11):5088-90.
1995 Commentary Archaea: narrowing the gap between prokaryotes and eukaryotes pdf
Information on Archaea:
Information on Archaea from the American Society for Microbiology
An Introduction to Archaea from the Department of Plant Biology at University of California, Berkeley
USGS Microbiology Laboratory, Columbus, Ohio
USGS Microbiology Research: Activity, Biogeochemistry, and Transport in Water
U.S. Environmental Protection Agency's Microbiology Home Page
U.S. Department of Energy's Microbial Genome Program
Archaea Taxonomy from the National Institutes of Health
News:
A hydrogen-based subsurface microbial community dominated by methanogens: Nature
Scientist May Have Discovered a Model for Extraterrestrial Life, Washington Post
Found: Life on Earth That Could Exist on Mars, New York Times (by Associated Press)
Scientists Find New Kind of Microbe, New York Times (by Associated Press)
UMass Researchers Find Environment on Earth that Mimics Mars Geochemically and Supports Ancient Life Form, NASA Press Release
Archaea can be distinguished from bacteria in that their cell walls do not have murein—a peptidoglycan-containing muramic acid. Their plasma membranes are ~4-5 nm in thickness, and are unique in including isopranyl ether lipids – branched isoprenoid hydrocarbons that are linked to glycerol by di-/tetraether bonds. The glycerol diethers are associated with bilayer membranes, whereas the glycerol tetraethers are associated with monolayer membranes. Archaea with mixtures of di- & tetraethers have mixed mono- & bilayer- membranes. Hyperthermophiles have increased membrane fluidity due to branched glycerol tetraethers, and often lack a cell wall. Psychrophiles have lipids comprising mainly unsaturated fatty acids. The plasma membrane of Archaea also contains sulfolipids.
The cell walls of most Archaea comprise a multilayered sacculus (sheet) that surrounds the plasma membrane and provides resistance to osmotic shock. This enables the Archaea (except Thermoplasma) to maintain high intracellular osmotic pressures (15-20 atm). The cells wall may comprise peptidoglycans (Methanobacterium) in which the chief sugars are N-acetyl talosaminouronic acid (NAT or T) and GlcNAc (NAG or G). These are linked to each other by lysozime insensitive (b1->3) glycosidic bond. The T units are linked to L-aminoacid containing oligopeptides by a penicillin-insensitive transpeptidase. In other genera, the cell wall may comprise only non-sulfated polysaccharides (Methanosarcina), only sulfated polysaccharides (Halococcus), only proteins (Methanomicrobium and Methanococcus), or negatively charged aminoacid-containing peptides (Halobacteria).
Images: tem -thermophilic archaeobacteria, line-up

Phylogenetic taxonomy of the Archaea (right), based on 16s rRNA trees.
The Bacteria (eubacteria) were the first prokaryotes recognized. (see phylogeny)
In 1977, Woese and Fox discovered the Archaea based upon their phylogenetic relationships to the Bacteria and the Eucarya. The yellow asterisk indicates the progenote, and the junction between teal and purple lines, the Last Common Ancestor, or Cenancestor.
Woese, Free Full Text Article: Toward a natural system of organisms
 (left) The Domain Archaea is divided into three Phyla: Crenarchaeota, Euryarchaeota, and Nanoarchaeota. A brief phylogenetic relationship of Crenarchaeota and Euryarchaeota are shown in the tree diagram (top right), and a more detailed cladogram is shown below. (click to enlarge)
(left) The Domain Archaea is divided into three Phyla: Crenarchaeota, Euryarchaeota, and Nanoarchaeota. A brief phylogenetic relationship of Crenarchaeota and Euryarchaeota are shown in the tree diagram (top right), and a more detailed cladogram is shown below. (click to enlarge)From the Taxonomicon, based on Systema Naturae, 2000:
Domain Archaea (Woese, Kandler & Wheelis, 1990)
Phylum Crenarchaeota
Class Thermoprotei
Order Thermoproteales™ Zillig & Stetter, 1982
Order Desulfurococcales
Order Sulfolobales Stetter, 1989
Phylum Euryarchaeota (Woese, Kandler & Wheelis, 1990)
Class Methanobacteria
Class Methanococci
Class Halobacteria
Class Thermoplasmata
Class Thermococci
Class Archaeoglobi Garrity & Holt, 2002
Class Methanopyri
Phylum Nanoarchaeota Huber et al., 2002
Genus Nanoarchaeum™ Huber et al., 2002
The Archaea: A Personal Overview of the Formative Years: "The simple method of growing methanogenic cells in an anaerobic pressurized atmosphere of hydrogen and carbon dioxide was pivotal to the discovery of the Archaea, for hydrogen and carbon dioxide could be replenished with safe containment of high levels of 32P and without exposure of cells to oxygen or contamination.
In November 1977, Woese and Fox proposed that ribosomal RNA sequence characterization could be used to define three "aboriginal lines of descent" (Woese and Fox, 1977). One line, the typical bacteria, was designated "eubacteria." "archaebacteria" was proposed as a name for the methanogen line, and the term "urkaryotes" was proposed for the cytoplasmic component of eukaryotic cells. So the methanogens were Archaebacteria. The name Archaebacteria was suggested by David Nanney. In a way, the term was unfortunate because the case was being made that the methanogens were "old" (i.e., represented a very ancient divergence in evolution), so that they were no more related to bacteria than to eukaryotes. On the other hand, they were bacteria. (Over a decade later, Woese, Kandler, and Wheelis would propose the name "Archaea" for the Archaebacteria [Woese et al., 1990]. By that time, much evidence had accumulated showing the Archaea clearly belonged on the eukaryotic line of descent, and it would be less confusing if the word "bacteria" in Archaebacteria was deleted.)"
more detail: Physiology of the Archaea :
Free Full Text Article : Phylogenetic structure of the prokaryotic domain: The primary kingdoms. Woese CR, Fox GE. Proc Natl Acad Sci U S A. 1977 Nov;74(11):5088-90.
1995 Commentary Archaea: narrowing the gap between prokaryotes and eukaryotes pdf
Information on Archaea:
Information on Archaea from the American Society for Microbiology
An Introduction to Archaea from the Department of Plant Biology at University of California, Berkeley
USGS Microbiology Laboratory, Columbus, Ohio
USGS Microbiology Research: Activity, Biogeochemistry, and Transport in Water
U.S. Environmental Protection Agency's Microbiology Home Page
U.S. Department of Energy's Microbial Genome Program
Archaea Taxonomy from the National Institutes of Health
News:
A hydrogen-based subsurface microbial community dominated by methanogens: Nature
Scientist May Have Discovered a Model for Extraterrestrial Life, Washington Post
Found: Life on Earth That Could Exist on Mars, New York Times (by Associated Press)
Scientists Find New Kind of Microbe, New York Times (by Associated Press)
UMass Researchers Find Environment on Earth that Mimics Mars Geochemically and Supports Ancient Life Form, NASA Press Release
Labels: Archaea, Archaeobacteria, cell walls, extremophiles, isopranyl ether lipids, Mendosicutes, NAG, NAT, phylogenetic taxonomy, sulfolipids
Interactions in Bacteria
Quorum sensing employs chemical signals for bacterial communication concerning achievement of critical mass. This process employs the production, release, and subsequent detection of chemical signaling molecules called autoinducers, and enables bacteria to regulate gene expression in response to changes in cell-population density. In response to quorum signals, bacteria alter in unison processes that are effective only when sufficient cells are available for coordinated activity. As bacterial populations grow, extracellular concentration of autoinducer increases until a threshold is reached at which the population responds with a population-wide alteration in gene expression. The processes controlled by quorum sensing are usually unproductive for an individual bacterium yet effective when undertaken by the group –bioluminescence, secretion of virulence factors, biofilm formation, sporulation, and the exchange of DNA.
Bacteria employ multiple chemical signaling molecules called autoinducers (AIs), one of which, AI-2, enables communication between other bacterial species in the vicinity. A gene called luxS is required for production of AI-2, and hundreds of species of bacteria have the luxS gene and employ AI-2 to communicate, suggesting that bacteria have a universal chemical language for communication between species. The LuxI protein synthesizes an acyl-homoserine lactone autoinducer, and the LuxR protein is responsible for autoinducer binding and subsequent activation of transcription of the luciferase operon. Cell-to-Cell Communication in Bacteria . Say What? Bacterial Conversation-Stoppers .
Cell-Cell Interactions in Bacteria : "One current estimate suggests that more than 99% of the bacteria on earth live as cell masses (Costerton et al., 1995), a condition conducive to cell interactions. As the broader significance of multicellular microbial life has been recognized, the cell interactions that facilitate multicellularity have been revealed."
"Now it is generally accepted that bacteria produce, and respond as groups to, chemical signals and that this interaction can lead to the coordination of group bacterial activities. This phenomenon has become known as quorum sensing (2-4). We also understand that groups of bacteria can form physical structures with unique characteristics, so-called biofilms (5, 6). Quorum sensing and biofilm biology have become very active areas in microbiology, and a large group of investigators is working on these fascinating aspects of bacterial biology, hoping to develop new therapeutic agents to treat associated persistent bacterial infections."
E. Peter Greenberg Bacterial communication and group behavior. J. Clin. Invest. 112:1288-1290 (2003). doi:10.1172/JCI200320099
"Quorum sensing uses signaling molecules, known as autoinducers [molecules that regulate mRNA production for specific genes in response to population density]. These are continuously produced by bacteria and can readily diffuse through the cell membrane. When elevated numbers of bacteria are present in an area, the concentration of autoinducers in the region will be higher. Autoinducer molecules (which include certain peptides and compounds known as homoserine lactones) can interact with specific repressor or activator sequences in DNA. The presence or absence of the autoinducer thus controls the production of mRNA, and therefore protein. These proteins are encoded by dozens of genes, including the genes for biofilm production. Laboratory strains of P. aeruginosa lacking the gene for a specific homoserine lactone will not develop into normal biofilms but pile up into a disorganized heap. From the bacteria's perspective, intracellular signaling has many advantages. Microbes often produce antibiotics that inhibit the growth of competitive species. Intracellular signaling not only brings bacteria together in biofilms, it also regulates the coordinated delivery of high doses of these antibiotics from the denser bacterial population. It also helps bacteria coordinate the release of virulence factors (such as disease-causing toxins) to overcome animal or plant defenses. Signals between bacteria in close proximity, as in a biofilm, also seem to enhance bacterial mating and the acquisition of novel DNA by transformation, both of which increase bacterial diversity." Biofilm Formation and Bacterial Communication.
"Now, the Kornberg team has found that P. aeruginosa bacteria without the gene [for PPK, polyphosphate kinase] are also unable to form microbial communities called biofilms, and they are unable to communicate with each other via a process called quorum sensing. Biofilms and quorum sensing are phenomena that occur when the bacteria encounter a new environment and must adapt to survive. Quorum-sensing communication between bacteria involves the release of small molecules that float among the bacteria and deliver a chemical message. When the bacteria determine, via quorum-sensing communication, that a critical mass of organisms has assembled, they use their flagella to slide toward each other and construct a thick, slimy biofilm." Bacterial communication, toxin production tied to intriguing cell protein, and:
"Polyphosphate is a long, chain-like molecule found in every living cell. Scientists believe that in animals, one of its roles may be to serve as a phosphate storage reservoir for the production of ATP (adenosine triphosphate), which provides the energy to power a cell. In bacteria, polyphosphate helps these single-celled organisms adapt to nutritional deficiencies and environmental stresses. It also helps them survive the state of suspended animation known as the stationary phase of growth. "
Bacteria also communicate via conjugation, which enables bacteria to exchange genetic material because of tube-like connections called pili. The donor bacterium contains conjugative or mobilizable genetic elements, usually a conjugative plasmid or episome plasmid that can integrate itself into the bacterial chromosome by genetic recombination. One such conjugative plasmid is called the F-plasmid. This is an episome about 100 thousand base-pairs in length, which carries its own origin of replication, called oriV. Most conjugative plasmids have systems ensuring that the recipient cell does not already contain a similar element, ensuring that there is only one copy of the F-plasmid in the F-positive bacterium. Genetic Construct for F and R cell types.
: New Salmonella Finding—Inter-Bacterial Communication! : Princeton - Bacterial communication :
Mahadevan, S (2002) Bacterial communication. Journal of Biosciences 27(5):pp. 443-444. Full text available as:PDF.
Phosphorylation switches
response regulator proteins in bacteria
Molecular Mechanisms of Signal Transduction : "The majority of bacterial response regulator proteins are transcription factors that serve as repressors or activators to regulate the expression of specific genes. The effector domains of these response regulators are DNA-binding domains that can be categorized into three major families based on sequence and structural similarity.
The OmpR/PhoB family of response regulator transcription factors, distinguished by a winged-helix DNA-binding domain, is the largest family, accounting for ~45 percent of all response regulators. Most characterized members of this family have been shown to bind as tandem dimers to direct repeat DNA recognition sequences. . . Phosphorylation induces dimerization or higher-order oligomerization of the proteins and that dimerization is mediated by the phosphorylated regulatory domains. The activated regulatory domains of Escherichia coli ArcA, KdpE, PhoB, PhoP, and TorR and T. maritima DrrB and DrrD all exist as dimers with identical alpha4-beta5-alpha5 interfaces. In all cases, the dimerization interface is formed by a few hydrophobic residues surrounded by an extensive network of intra- and intermolecular salt bridges. The residues involved in these interactions are highly conserved in all members of the OmpR/PhoB family, but not in other response regulators. It is proposed that this mode of dimerization is common to most members of the OmpR/PhoB family and that it represents a family-specific mechanism for activation of DNA binding. Upon phosphorylation, the interface between regulatory and DNA-binding domains is disrupted, allowing the regulatory domains to dimerize via their alpha4-beta5-aalpha5 faces. Disruption of the interdomain interface frees the DNA-binding domain, allowing it to dimerize in tandem on direct repeat DNA half-sites with symmetry that is different from that of the regulatory domain dimer. Additional nuclear magnetic resonance (NMR) and biochemical studies support this mechanism of activation. " Link to link-to-images.
Bacteria employ multiple chemical signaling molecules called autoinducers (AIs), one of which, AI-2, enables communication between other bacterial species in the vicinity. A gene called luxS is required for production of AI-2, and hundreds of species of bacteria have the luxS gene and employ AI-2 to communicate, suggesting that bacteria have a universal chemical language for communication between species. The LuxI protein synthesizes an acyl-homoserine lactone autoinducer, and the LuxR protein is responsible for autoinducer binding and subsequent activation of transcription of the luciferase operon. Cell-to-Cell Communication in Bacteria . Say What? Bacterial Conversation-Stoppers .
Cell-Cell Interactions in Bacteria : "One current estimate suggests that more than 99% of the bacteria on earth live as cell masses (Costerton et al., 1995), a condition conducive to cell interactions. As the broader significance of multicellular microbial life has been recognized, the cell interactions that facilitate multicellularity have been revealed."
"Now it is generally accepted that bacteria produce, and respond as groups to, chemical signals and that this interaction can lead to the coordination of group bacterial activities. This phenomenon has become known as quorum sensing (2-4). We also understand that groups of bacteria can form physical structures with unique characteristics, so-called biofilms (5, 6). Quorum sensing and biofilm biology have become very active areas in microbiology, and a large group of investigators is working on these fascinating aspects of bacterial biology, hoping to develop new therapeutic agents to treat associated persistent bacterial infections."
E. Peter Greenberg Bacterial communication and group behavior. J. Clin. Invest. 112:1288-1290 (2003). doi:10.1172/JCI200320099
"Quorum sensing uses signaling molecules, known as autoinducers [molecules that regulate mRNA production for specific genes in response to population density]. These are continuously produced by bacteria and can readily diffuse through the cell membrane. When elevated numbers of bacteria are present in an area, the concentration of autoinducers in the region will be higher. Autoinducer molecules (which include certain peptides and compounds known as homoserine lactones) can interact with specific repressor or activator sequences in DNA. The presence or absence of the autoinducer thus controls the production of mRNA, and therefore protein. These proteins are encoded by dozens of genes, including the genes for biofilm production. Laboratory strains of P. aeruginosa lacking the gene for a specific homoserine lactone will not develop into normal biofilms but pile up into a disorganized heap. From the bacteria's perspective, intracellular signaling has many advantages. Microbes often produce antibiotics that inhibit the growth of competitive species. Intracellular signaling not only brings bacteria together in biofilms, it also regulates the coordinated delivery of high doses of these antibiotics from the denser bacterial population. It also helps bacteria coordinate the release of virulence factors (such as disease-causing toxins) to overcome animal or plant defenses. Signals between bacteria in close proximity, as in a biofilm, also seem to enhance bacterial mating and the acquisition of novel DNA by transformation, both of which increase bacterial diversity." Biofilm Formation and Bacterial Communication.
"Now, the Kornberg team has found that P. aeruginosa bacteria without the gene [for PPK, polyphosphate kinase] are also unable to form microbial communities called biofilms, and they are unable to communicate with each other via a process called quorum sensing. Biofilms and quorum sensing are phenomena that occur when the bacteria encounter a new environment and must adapt to survive. Quorum-sensing communication between bacteria involves the release of small molecules that float among the bacteria and deliver a chemical message. When the bacteria determine, via quorum-sensing communication, that a critical mass of organisms has assembled, they use their flagella to slide toward each other and construct a thick, slimy biofilm." Bacterial communication, toxin production tied to intriguing cell protein, and:
"Polyphosphate is a long, chain-like molecule found in every living cell. Scientists believe that in animals, one of its roles may be to serve as a phosphate storage reservoir for the production of ATP (adenosine triphosphate), which provides the energy to power a cell. In bacteria, polyphosphate helps these single-celled organisms adapt to nutritional deficiencies and environmental stresses. It also helps them survive the state of suspended animation known as the stationary phase of growth. "
Bacteria also communicate via conjugation, which enables bacteria to exchange genetic material because of tube-like connections called pili. The donor bacterium contains conjugative or mobilizable genetic elements, usually a conjugative plasmid or episome plasmid that can integrate itself into the bacterial chromosome by genetic recombination. One such conjugative plasmid is called the F-plasmid. This is an episome about 100 thousand base-pairs in length, which carries its own origin of replication, called oriV. Most conjugative plasmids have systems ensuring that the recipient cell does not already contain a similar element, ensuring that there is only one copy of the F-plasmid in the F-positive bacterium. Genetic Construct for F and R cell types.
: New Salmonella Finding—Inter-Bacterial Communication! : Princeton - Bacterial communication :
Mahadevan, S (2002) Bacterial communication. Journal of Biosciences 27(5):pp. 443-444. Full text available as:PDF.
Phosphorylation switches
response regulator proteins in bacteria
Molecular Mechanisms of Signal Transduction : "The majority of bacterial response regulator proteins are transcription factors that serve as repressors or activators to regulate the expression of specific genes. The effector domains of these response regulators are DNA-binding domains that can be categorized into three major families based on sequence and structural similarity.
The OmpR/PhoB family of response regulator transcription factors, distinguished by a winged-helix DNA-binding domain, is the largest family, accounting for ~45 percent of all response regulators. Most characterized members of this family have been shown to bind as tandem dimers to direct repeat DNA recognition sequences. . . Phosphorylation induces dimerization or higher-order oligomerization of the proteins and that dimerization is mediated by the phosphorylated regulatory domains. The activated regulatory domains of Escherichia coli ArcA, KdpE, PhoB, PhoP, and TorR and T. maritima DrrB and DrrD all exist as dimers with identical alpha4-beta5-alpha5 interfaces. In all cases, the dimerization interface is formed by a few hydrophobic residues surrounded by an extensive network of intra- and intermolecular salt bridges. The residues involved in these interactions are highly conserved in all members of the OmpR/PhoB family, but not in other response regulators. It is proposed that this mode of dimerization is common to most members of the OmpR/PhoB family and that it represents a family-specific mechanism for activation of DNA binding. Upon phosphorylation, the interface between regulatory and DNA-binding domains is disrupted, allowing the regulatory domains to dimerize via their alpha4-beta5-aalpha5 faces. Disruption of the interdomain interface frees the DNA-binding domain, allowing it to dimerize in tandem on direct repeat DNA half-sites with symmetry that is different from that of the regulatory domain dimer. Additional nuclear magnetic resonance (NMR) and biochemical studies support this mechanism of activation. " Link to link-to-images.
Bacterial infection
The invasion of an organism by a bacterial pathogen is called infection. Bacteria themselves can be infected by viral bacteriophages (bacterium eaters).
Bacterial motility
Many prokaryotes are not motile, however motile prokaryotes employ a number of mechanisms:
1. Flagella - Gram -ve, Gram +ve
2. Gliding
3. Buoyancy change
4. Axial filaments - Spirochetes,
Prokaryotic movement is controlled by chemotactic signaling (below)
Bacterial flagella are rigid, hollow structures constructed of the protein flagellin (FliC). Each long filament is attached by a hook to the basal body, which serves as the motor.
The filament is usually about 20 nm in diameter and typically comprises of thousands of copies of a single form of flagellin. Less often the filament is composed of several different flagellins. A capping protein, HAP2, is located at the tip of the flagellum. The junction of the single-protein hook and filament requires hook-associated proteins called HAP1 and HAP3. The basal bodies of Gram +ve bacteria are anchored in the plasma membrane. The basal structure comprises a rod, a series of rings, the Mot proteins, the switch complex and the flagellum-specific export apparatus. The rings anchor the flagellum to the cytoplasmic membrane (MS ring), the peptidoglycan (P ring) and the outer membrane (L ring). The Gram -ve bacteria lack the P and L rings, and instead have an additional basal body embedded in the outer membrane. The switch proteins (FliG, FliM and FliN) permit the flagellum to switch rotation, thus controlling the direction of swimming in response to attractants or repellents in the environment (chemotaxis system). As a result of environmental sensing, phosphorylated CheY protein comes into direct contact with the FliM switch protein (Bourret et al., 2002). MotA and MotB proteins form a channel through which flow the protons that power the rotation of the flagellum. They form the stator, or nonrotating portion, of the structure where MotB is apparently attached to the peptidoglycan layer. The rotor extends into the cytoplasm (forming the C ring) and comprising several proteins including the three switch proteins. Image with legend : Diagram : image bacterial flagellar nanomotor : diagram :
Note: the bacterial flagellum is the target of intellectualized creationist nonsense in the guise of claims of so-called "irreducible complexity", which take the position that assemblages of the individual components of molecular machinery could not have evolved, but could only be explained by design (God). For refutation see: Research has demonstrated that assembly of pre-existing modifications operate in subsequently evolved features.* Reducible complexity : "As an icon of anti-evolution, the flagellum has fallen." The Flagellum unspun : Flagellum evolution in Nature Reviews Microbiology re Pallen MJ, Matzke NJ. (2006). “From The Origin of Species to the origin of bacterial flagella.” Nature Reviews Microbiology, 4(10), 784-790. October 2006. Advanced Online Publication on September 5, 2006. [PubMed] [Journal] [DOI] [Google Scholar] : Evolution in (Brownian) space: a model for the origin of the bacterial flagellum & Background to "Evolution in (Brownian) space: a model for the origin of the bacterial flagellum" : Inventing the dynamo machine: the evolution of the F-type and V-type ATPases :
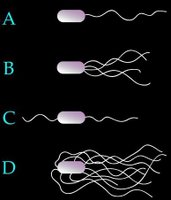 Arrangement of eubacterial flagella
Arrangement of eubacterial flagella
A. monotrichous – polar – Pseudomonas sp.,
B. lophotrichous – Spirillum
C. amphitrichous –
D. peritrichous – Proteus vulgaris, sem - peritrichous flagella, E. coli,
Mechanisms of flagellar motility in eubacteria:
Motile behavior of bacteria : Sense and sensibility in bacteria : Animated motor :
Archaeal flagella are superficially similar to those of eubacteria, but are not homologous to bacterial flagella because of a number of differences in structure, power source, and growth. [1, 2, 3, 4] Diagram - Archaeal flagellum :
The Archaeal flagellum is:
1. only 15 nm in diameter, rather than 20 nm as in eubacteria.
2. appears to grow at base rather than tip
3. is homologous to Type IV pili, unlike eubacteria
4. is coded by genes without sequence homology to the eubacteria
The archaeal flagellum: a different kind of prokaryotic motility structure.
The archaeal flagellum is a unique motility apparatus distinct in composition and likely in assembly from the bacterial flagellum. Gene families comprised of multiple flagellin genes co-transcribed with a number of conserved, archaeal-specific accessory genes have been identified in several archaea. However, no homologues of any bacterial genes involved in flagella structure have yet been identified in any archaeon, including those archaea in which the complete genome sequence has been published. Archaeal flagellins possess a highly conserved hydrophobic N-terminal sequence that is similar to that of type IV pilins and clearly unlike that of bacterial flagellins. Also unlike bacterial flagellins but similar to type IV pilins, archaeal flagellins are initially synthesized with a short leader peptide that is cleaved by a membrane-located peptidase. Thomas NA, Bardy SL, Jarrell KF. The archaeal flagellum: a different kind of prokaryotic motility structure. FEMS Microbiol Rev. 2001 Apr;25(2):147-74.
Recent advances in the structure and assembly of the archaeal flagellum. [J Mol Microbiol Biotechnol. 2004] PMID: 15170402
Characterization of flagellum gene families of methanogenic archaea and localization of novel flagellum accessory proteins. [J Bacteriol. 2001] PMID: 11717274
Prokaryotic motility structures. [Microbiology. 2003] PMID: 12624192
Mutants in flaI and flaJ of the archaeon Methanococcus voltae are deficient in flagellum assembly. [Mol Microbiol. 2002] PMID: 12410843
Stress genes and proteins in the archaea. [Microbiol Mol Biol Rev. 1999]
Both eubacterial and archaeal flagella differ from the flagella of eukaryotes:
Eukaryotic cilia and flagella both have an internal structure built upon microtubules, but the flagellum is longer and is more often a single organelle. Inside both cilia and flagella is a microtubule-based cytoskeleton termed the axoneme, which provides scaffolding for various protein complexes.
Chemotaxis is a signal transduction systems that controls movement. Even though the motility apparatus differs among organisms, the general mechanism of control of chemotaxis is conserved throughout all bacteria and archaea. The chemotaxis mechanism in B. subtilis is probably close to that of the ancestral organism from which the bacteria and archaea descended. The chemotactic system of E. coli is streamlined and lacks, or has significantly modified, some basic features of the primordial mechanism that existed when the bacteria and archaea separated during biological evolution.
Central to chemotactic control is the two-component system in which phosphorylation of a response regulator reflects phosphorylation of a histidine autokinase that senses environmental parameters (117). This is the commonest mode of signal transduction system in bacteria, and the "two component system" controls diverse processes such as gene expression, sporulation, and chemotaxis. • Phosphorylation switches •
In prokaryotes, most signaling is effected by a simple two-component systems (TCS), which may bring about changes in gene transcription or chemotactic (swimming) behavior of the organism. In bacteria, most TCS are produced by one histidine kinase (HK) and its corresponding response regulator (RR). After detection of a signal, such as an alteration in the cell's redox status or the ion concentration in the medium, two HK monomers dimerize and phosphorylate a histidine residue in trans. Subsequently the phosphate group is transferred to an aspartate residue of the RR. This short phosphorylation cascade (figure showing basic two-component phosphotransfer scheme) can impact chemotaxis (via the CheA-CheY-system) and the pattern of gene expression. At the receptors, chemotactic signals control autophosphorylation of the CheA histidine kinase. The phosphohistidine acts as a substrate for the response regulator CheY, which catalyzes the transfer of the phosphoryl group to a conserved aspartate (review 250). The resulting phosphorylated CheY-P can interact with the switch mechanism in the motor (42, 149, 186, 193, 234), causing a change in movement, such as in direction or speed of rotation of flagella.
Chemotaxis proteins comprise four groups—a signal recognition and transduction group, an excitation group, an adaptation group, and a signal removal group (to dephosphorylate CheY-P). The signal recognition and transduction group includes the receptors (9, 81, 118) and ligand binding proteins (4, 86), which are capable of binding effectors outside the cell; a few receptors, however, are cytoplasmic (92, 93, 229). Image general chemotaxis model : Table proteins in chemotaxis :
Some prokaryotic TCSs are more sophisticated than the basic HK-RR system. These comprise a "hybridkinase", which consists of a kinase domain and a response regulator domain plus an additional phosphorylatable histidine residue. These modified TCS are also called "phosphorelays". Because two or more phosphorylation events (figure showing phosphorelay system) occur, it is believed that these systems can integrate more signals into the signaling cascade and are more fine tuned. Prokaryotic phosphorelays are a minority in the TCS-family, whereas they are the major type of two-component systems found in eukaryotes. Increasingly, TCSs have been demonstrated to cross-communicate (figure showing cross talk between the Pho- and Pmr-TCS in Salmonella enterica), enhancing the organism's responsiveness to the environment.
Abstracts of Articles:
Diversity in chemotaxis mechanisms among the bacteria and archaea.
The study of chemotaxis describes the cellular processes that control the movement of organisms toward favorable environments. In bacteria and archaea, motility is controlled by a two-component system involving a histidine kinase that senses the environment and a response regulator, a very common type of signal transduction in prokaryotes. Most insights into the processes involved have come from studies of Escherichia coli over the last three decades. However, in the last 10 years, with the sequencing of many prokaryotic genomes, it has become clear that E. coli represents a streamlined example of bacterial chemotaxis. While general features of excitation remain conserved among bacteria and archaea, specific features, such as adaptational processes and hydrolysis of the intracellular signal CheY-P, are quite diverse. The Bacillus subtilis chemotaxis system is considerably more complex and appears to be similar to the one that existed when the bacteria and archaea separated during evolution, so that understanding this mechanism should provide insight into the variety of mechanisms used today by the broad sweep of chemotactic bacteria and archaea. However, processes even beyond those used in E. coli and B. subtilis have been discovered in other organisms. This review emphasizes those used by B. subtilis and these other organisms but also gives an account of the mechanism in E. coli.
Szurmant H, Ordal GW. Diversity in Chemotaxis Mechanisms among the Bacteria and Archaea. (Free Full Text Article) Microbiol Mol Biol Rev. 2004 Jun;68(2):301-19.
Prokaryotic motility structures.
Prokaryotes use a wide variety of structures to facilitate motility. The majority of research to date has focused on swimming motility and the molecular architecture of the bacterial flagellum. While intriguing questions remain, especially concerning the specialized export system involved in flagellum assembly, for the most part the structural components and their location within the flagellum and function are now known. The same cannot be said of the other apparati including archaeal flagella, type IV pili, the junctional pore, ratchet structure and the contractile cytoskeleton used by a variety of organisms for motility. In these cases, many of the structural components have yet to be identified and the mechanism of action that results in motility is often still poorly understood. Research on the bacterial flagellum has greatly aided our understanding of not only motility but also protein secretion and genetic regulation systems. Continued study and understanding of all prokaryotic motility structures will provide a wealth of knowledge that is sure to extend beyond the bounds of prokaryotic movement.
Bardy SL, Ng SY, Jarrell KF. Prokaryotic motility structures. (Free Full Text Article) Microbiology. 2003 Feb;149(Pt 2):295-304.
The archaeal flagellum: a different kind of prokaryotic motility structure. [FEMS Microbiol Rev. 2001] PMID: 11250034
Recent advances in the structure and assembly of the archaeal flagellum. [J Mol Microbiol Biotechnol. 2004] PMID: 15170402
Type II protein secretion and its relationship to bacterial type IV pili and archaeal flagella. [Microbiology. 2003] PMID: 14600218
Diversity in chemotaxis mechanisms among the bacteria and archaea. [Microbiol Mol Biol Rev. 2004] PMID: 15187186
Bacterial and archaeal flagella as prokaryotic motility organelles. [Biochemistry (Mosc). 2004] PMID: 15627373
See all Related Articles...
Characterization of flagellum gene families of methanogenic archaea and localization of novel flagellum accessory proteins. (Free Full Text Article) Thomas NA, Jarrell KF. J Bacteriol. 2001 Dec;183(24):7154-64.
Bourret,R. B., Charon, N. W., Stock, A. M. & West, A. H. (2002). Bright lights, abundant operons – fluorescence and genomic technologies advance studies of bacterial locomotion and signal transduction: review of the BLAST meeting, Cuernavaca, Mexico, 14–19 January 2001. J Bacteriol 184, 1–17. [Free Full Text]
3D diagram - axoneme : 3D animation – inside flagellum : image - detail of cilia : tem - structure cilium : diagram - mechanism of ciliary motility : Geometric Clutch Model : animation - cilia & flagella
1. Flagella - Gram -ve, Gram +ve
2. Gliding
3. Buoyancy change
4. Axial filaments - Spirochetes,
Prokaryotic movement is controlled by chemotactic signaling (below)
Bacterial flagella are rigid, hollow structures constructed of the protein flagellin (FliC). Each long filament is attached by a hook to the basal body, which serves as the motor.
The filament is usually about 20 nm in diameter and typically comprises of thousands of copies of a single form of flagellin. Less often the filament is composed of several different flagellins. A capping protein, HAP2, is located at the tip of the flagellum. The junction of the single-protein hook and filament requires hook-associated proteins called HAP1 and HAP3. The basal bodies of Gram +ve bacteria are anchored in the plasma membrane. The basal structure comprises a rod, a series of rings, the Mot proteins, the switch complex and the flagellum-specific export apparatus. The rings anchor the flagellum to the cytoplasmic membrane (MS ring), the peptidoglycan (P ring) and the outer membrane (L ring). The Gram -ve bacteria lack the P and L rings, and instead have an additional basal body embedded in the outer membrane. The switch proteins (FliG, FliM and FliN) permit the flagellum to switch rotation, thus controlling the direction of swimming in response to attractants or repellents in the environment (chemotaxis system). As a result of environmental sensing, phosphorylated CheY protein comes into direct contact with the FliM switch protein (Bourret et al., 2002). MotA and MotB proteins form a channel through which flow the protons that power the rotation of the flagellum. They form the stator, or nonrotating portion, of the structure where MotB is apparently attached to the peptidoglycan layer. The rotor extends into the cytoplasm (forming the C ring) and comprising several proteins including the three switch proteins. Image with legend : Diagram : image bacterial flagellar nanomotor : diagram :
Note: the bacterial flagellum is the target of intellectualized creationist nonsense in the guise of claims of so-called "irreducible complexity", which take the position that assemblages of the individual components of molecular machinery could not have evolved, but could only be explained by design (God). For refutation see: Research has demonstrated that assembly of pre-existing modifications operate in subsequently evolved features.* Reducible complexity : "As an icon of anti-evolution, the flagellum has fallen." The Flagellum unspun : Flagellum evolution in Nature Reviews Microbiology re Pallen MJ, Matzke NJ. (2006). “From The Origin of Species to the origin of bacterial flagella.” Nature Reviews Microbiology, 4(10), 784-790. October 2006. Advanced Online Publication on September 5, 2006. [PubMed] [Journal] [DOI] [Google Scholar] : Evolution in (Brownian) space: a model for the origin of the bacterial flagellum & Background to "Evolution in (Brownian) space: a model for the origin of the bacterial flagellum" : Inventing the dynamo machine: the evolution of the F-type and V-type ATPases :
 Arrangement of eubacterial flagella
Arrangement of eubacterial flagellaA. monotrichous – polar – Pseudomonas sp.,
B. lophotrichous – Spirillum
C. amphitrichous –
D. peritrichous – Proteus vulgaris, sem - peritrichous flagella, E. coli,
Mechanisms of flagellar motility in eubacteria:
Motile behavior of bacteria : Sense and sensibility in bacteria : Animated motor :
Archaeal flagella are superficially similar to those of eubacteria, but are not homologous to bacterial flagella because of a number of differences in structure, power source, and growth. [1, 2, 3, 4] Diagram - Archaeal flagellum :
The Archaeal flagellum is:
1. only 15 nm in diameter, rather than 20 nm as in eubacteria.
2. appears to grow at base rather than tip
3. is homologous to Type IV pili, unlike eubacteria
4. is coded by genes without sequence homology to the eubacteria
The archaeal flagellum: a different kind of prokaryotic motility structure.
The archaeal flagellum is a unique motility apparatus distinct in composition and likely in assembly from the bacterial flagellum. Gene families comprised of multiple flagellin genes co-transcribed with a number of conserved, archaeal-specific accessory genes have been identified in several archaea. However, no homologues of any bacterial genes involved in flagella structure have yet been identified in any archaeon, including those archaea in which the complete genome sequence has been published. Archaeal flagellins possess a highly conserved hydrophobic N-terminal sequence that is similar to that of type IV pilins and clearly unlike that of bacterial flagellins. Also unlike bacterial flagellins but similar to type IV pilins, archaeal flagellins are initially synthesized with a short leader peptide that is cleaved by a membrane-located peptidase. Thomas NA, Bardy SL, Jarrell KF. The archaeal flagellum: a different kind of prokaryotic motility structure. FEMS Microbiol Rev. 2001 Apr;25(2):147-74.
Recent advances in the structure and assembly of the archaeal flagellum. [J Mol Microbiol Biotechnol. 2004] PMID: 15170402
Characterization of flagellum gene families of methanogenic archaea and localization of novel flagellum accessory proteins. [J Bacteriol. 2001] PMID: 11717274
Prokaryotic motility structures. [Microbiology. 2003] PMID: 12624192
Mutants in flaI and flaJ of the archaeon Methanococcus voltae are deficient in flagellum assembly. [Mol Microbiol. 2002] PMID: 12410843
Stress genes and proteins in the archaea. [Microbiol Mol Biol Rev. 1999]
Both eubacterial and archaeal flagella differ from the flagella of eukaryotes:
Eukaryotic cilia and flagella both have an internal structure built upon microtubules, but the flagellum is longer and is more often a single organelle. Inside both cilia and flagella is a microtubule-based cytoskeleton termed the axoneme, which provides scaffolding for various protein complexes.
Chemotaxis is a signal transduction systems that controls movement. Even though the motility apparatus differs among organisms, the general mechanism of control of chemotaxis is conserved throughout all bacteria and archaea. The chemotaxis mechanism in B. subtilis is probably close to that of the ancestral organism from which the bacteria and archaea descended. The chemotactic system of E. coli is streamlined and lacks, or has significantly modified, some basic features of the primordial mechanism that existed when the bacteria and archaea separated during biological evolution.
Central to chemotactic control is the two-component system in which phosphorylation of a response regulator reflects phosphorylation of a histidine autokinase that senses environmental parameters (117). This is the commonest mode of signal transduction system in bacteria, and the "two component system" controls diverse processes such as gene expression, sporulation, and chemotaxis. • Phosphorylation switches •
In prokaryotes, most signaling is effected by a simple two-component systems (TCS), which may bring about changes in gene transcription or chemotactic (swimming) behavior of the organism. In bacteria, most TCS are produced by one histidine kinase (HK) and its corresponding response regulator (RR). After detection of a signal, such as an alteration in the cell's redox status or the ion concentration in the medium, two HK monomers dimerize and phosphorylate a histidine residue in trans. Subsequently the phosphate group is transferred to an aspartate residue of the RR. This short phosphorylation cascade (figure showing basic two-component phosphotransfer scheme) can impact chemotaxis (via the CheA-CheY-system) and the pattern of gene expression. At the receptors, chemotactic signals control autophosphorylation of the CheA histidine kinase. The phosphohistidine acts as a substrate for the response regulator CheY, which catalyzes the transfer of the phosphoryl group to a conserved aspartate (review 250). The resulting phosphorylated CheY-P can interact with the switch mechanism in the motor (42, 149, 186, 193, 234), causing a change in movement, such as in direction or speed of rotation of flagella.
Chemotaxis proteins comprise four groups—a signal recognition and transduction group, an excitation group, an adaptation group, and a signal removal group (to dephosphorylate CheY-P). The signal recognition and transduction group includes the receptors (9, 81, 118) and ligand binding proteins (4, 86), which are capable of binding effectors outside the cell; a few receptors, however, are cytoplasmic (92, 93, 229). Image general chemotaxis model : Table proteins in chemotaxis :
Some prokaryotic TCSs are more sophisticated than the basic HK-RR system. These comprise a "hybridkinase", which consists of a kinase domain and a response regulator domain plus an additional phosphorylatable histidine residue. These modified TCS are also called "phosphorelays". Because two or more phosphorylation events (figure showing phosphorelay system) occur, it is believed that these systems can integrate more signals into the signaling cascade and are more fine tuned. Prokaryotic phosphorelays are a minority in the TCS-family, whereas they are the major type of two-component systems found in eukaryotes. Increasingly, TCSs have been demonstrated to cross-communicate (figure showing cross talk between the Pho- and Pmr-TCS in Salmonella enterica), enhancing the organism's responsiveness to the environment.
Abstracts of Articles:
Diversity in chemotaxis mechanisms among the bacteria and archaea.
The study of chemotaxis describes the cellular processes that control the movement of organisms toward favorable environments. In bacteria and archaea, motility is controlled by a two-component system involving a histidine kinase that senses the environment and a response regulator, a very common type of signal transduction in prokaryotes. Most insights into the processes involved have come from studies of Escherichia coli over the last three decades. However, in the last 10 years, with the sequencing of many prokaryotic genomes, it has become clear that E. coli represents a streamlined example of bacterial chemotaxis. While general features of excitation remain conserved among bacteria and archaea, specific features, such as adaptational processes and hydrolysis of the intracellular signal CheY-P, are quite diverse. The Bacillus subtilis chemotaxis system is considerably more complex and appears to be similar to the one that existed when the bacteria and archaea separated during evolution, so that understanding this mechanism should provide insight into the variety of mechanisms used today by the broad sweep of chemotactic bacteria and archaea. However, processes even beyond those used in E. coli and B. subtilis have been discovered in other organisms. This review emphasizes those used by B. subtilis and these other organisms but also gives an account of the mechanism in E. coli.
Szurmant H, Ordal GW. Diversity in Chemotaxis Mechanisms among the Bacteria and Archaea. (Free Full Text Article) Microbiol Mol Biol Rev. 2004 Jun;68(2):301-19.
Prokaryotic motility structures.
Prokaryotes use a wide variety of structures to facilitate motility. The majority of research to date has focused on swimming motility and the molecular architecture of the bacterial flagellum. While intriguing questions remain, especially concerning the specialized export system involved in flagellum assembly, for the most part the structural components and their location within the flagellum and function are now known. The same cannot be said of the other apparati including archaeal flagella, type IV pili, the junctional pore, ratchet structure and the contractile cytoskeleton used by a variety of organisms for motility. In these cases, many of the structural components have yet to be identified and the mechanism of action that results in motility is often still poorly understood. Research on the bacterial flagellum has greatly aided our understanding of not only motility but also protein secretion and genetic regulation systems. Continued study and understanding of all prokaryotic motility structures will provide a wealth of knowledge that is sure to extend beyond the bounds of prokaryotic movement.
Bardy SL, Ng SY, Jarrell KF. Prokaryotic motility structures. (Free Full Text Article) Microbiology. 2003 Feb;149(Pt 2):295-304.
The archaeal flagellum: a different kind of prokaryotic motility structure. [FEMS Microbiol Rev. 2001] PMID: 11250034
Recent advances in the structure and assembly of the archaeal flagellum. [J Mol Microbiol Biotechnol. 2004] PMID: 15170402
Type II protein secretion and its relationship to bacterial type IV pili and archaeal flagella. [Microbiology. 2003] PMID: 14600218
Diversity in chemotaxis mechanisms among the bacteria and archaea. [Microbiol Mol Biol Rev. 2004] PMID: 15187186
Bacterial and archaeal flagella as prokaryotic motility organelles. [Biochemistry (Mosc). 2004] PMID: 15627373
See all Related Articles...
Characterization of flagellum gene families of methanogenic archaea and localization of novel flagellum accessory proteins. (Free Full Text Article) Thomas NA, Jarrell KF. J Bacteriol. 2001 Dec;183(24):7154-64.
Bourret,R. B., Charon, N. W., Stock, A. M. & West, A. H. (2002). Bright lights, abundant operons – fluorescence and genomic technologies advance studies of bacterial locomotion and signal transduction: review of the BLAST meeting, Cuernavaca, Mexico, 14–19 January 2001. J Bacteriol 184, 1–17. [Free Full Text]
3D diagram - axoneme : 3D animation – inside flagellum : image - detail of cilia : tem - structure cilium : diagram - mechanism of ciliary motility : Geometric Clutch Model : animation - cilia & flagella
Labels: Archaea, bacterial flagellum, bacterial motility, chemotactic signaling, chemotaxis, Gram stain, kinase, peptidoglycan, two-componets systems





































